Data modeling, annotation, and discovery
We want FAIRly managed data:
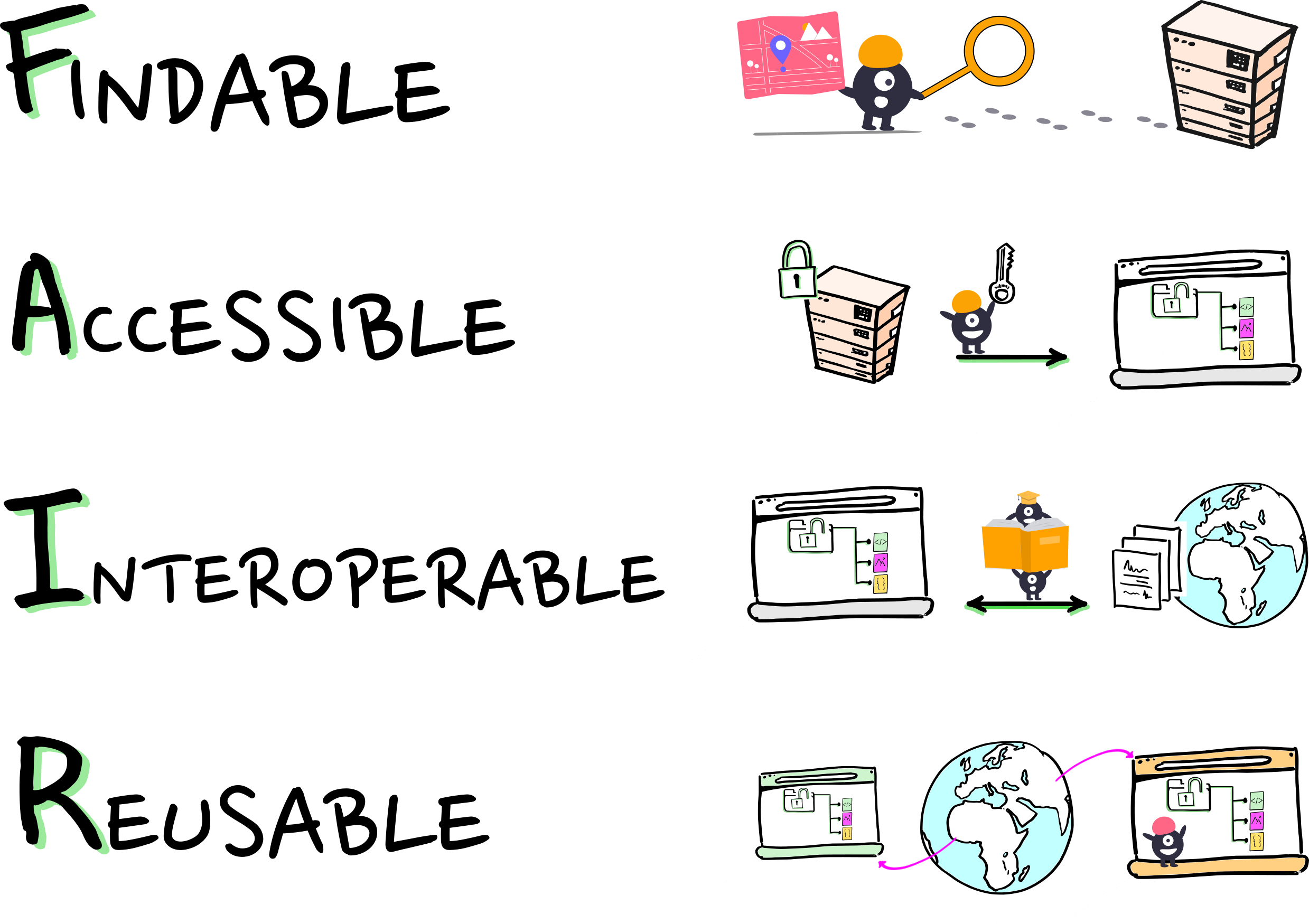 Wilkinson, et al. 2016. The FAIR Guiding Principles for scientific data management and stewardship. https://doi.org/10.1038/sdata.2016.18
Wilkinson, et al. 2016. The FAIR Guiding Principles for scientific data management and stewardship. https://doi.org/10.1038/sdata.2016.18
We want FAIRly managed data:
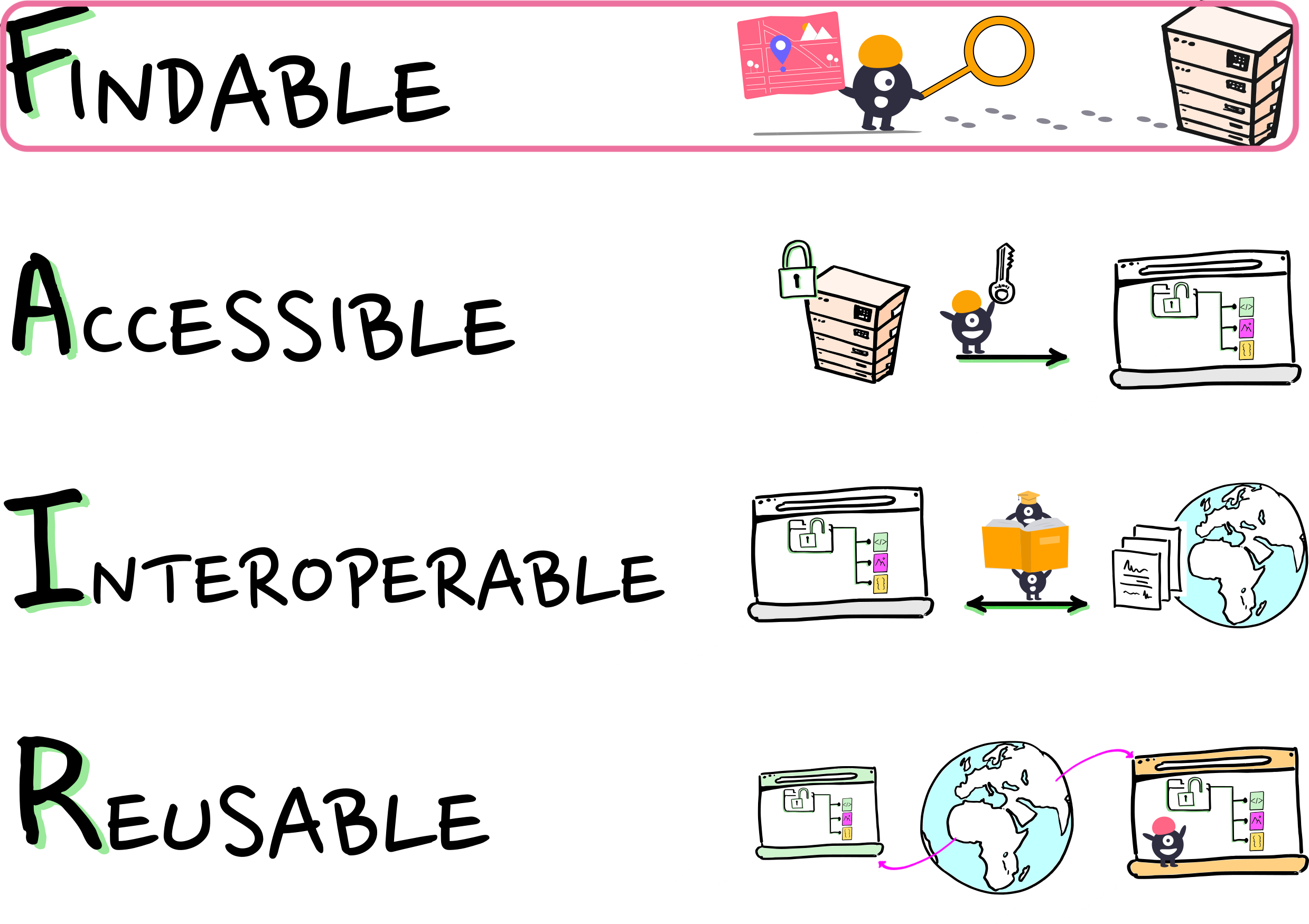 Wilkinson, et al. 2016. The FAIR Guiding Principles for scientific data management and stewardship. https://doi.org/10.1038/sdata.2016.18
Wilkinson, et al. 2016. The FAIR Guiding Principles for scientific data management and stewardship. https://doi.org/10.1038/sdata.2016.18
If everything has an identifier...
❱ cat .datalad/config
[datalad "dataset"]
id = fd3b41bb-5d75-4cee-b41c-aac0e0cae7f1❱ git log
commit 8b778413e195d5b91d7039e8e035fd788b4b25f5 (HEAD -> main, dhub/main)
Merge: e428b3a e9c8da7
Author: Stephan Heunis jsheunis@noreply.localhost>
Date: Tue Jun 24 12:00:44 2025 +0000
Merge pull request 'Add readme and license info' (#5) from readme-license into main
Reviewed-on: https://hub.datalad.org/edu/penguins/pulls/5
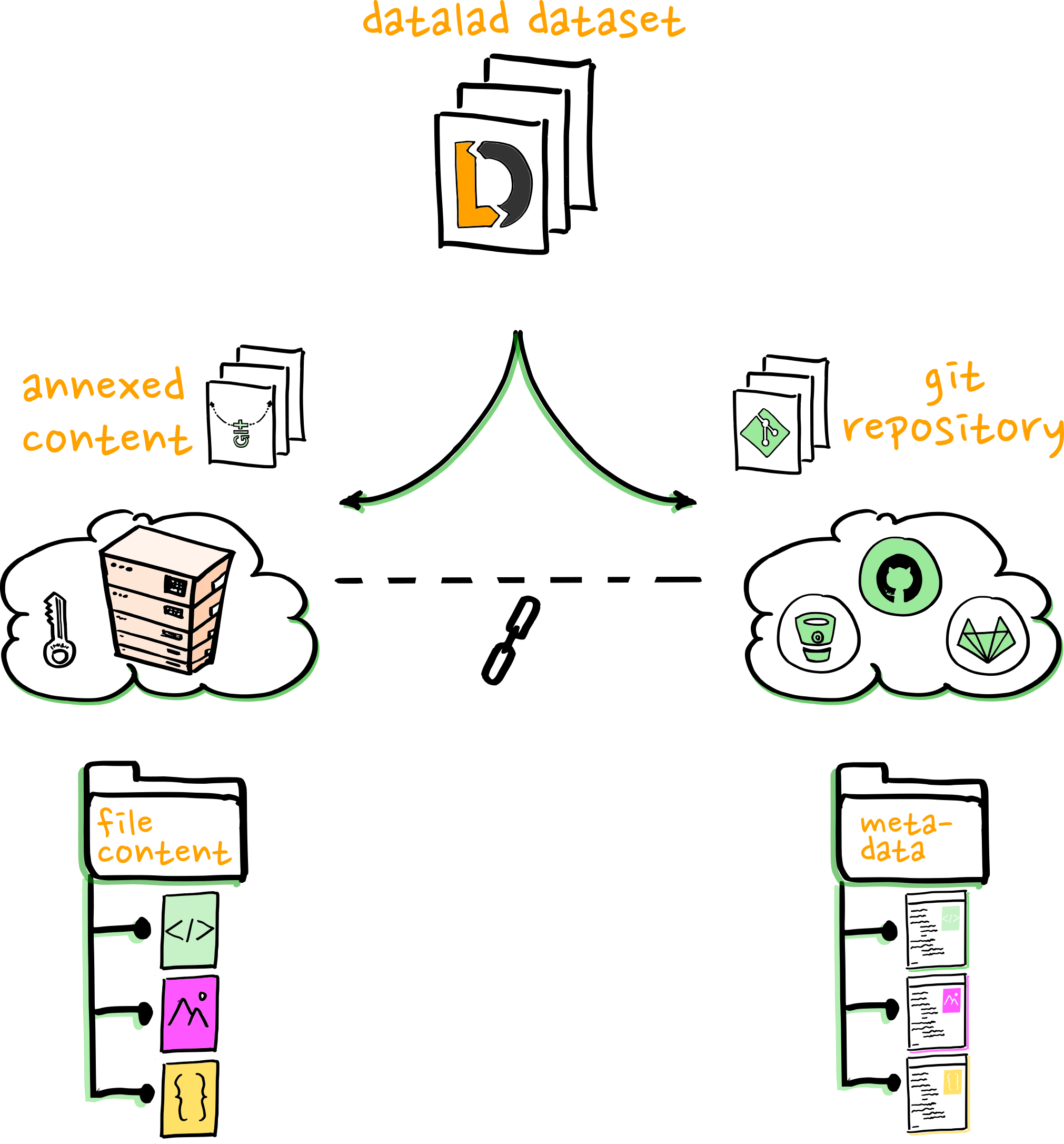
❱ git annex info adelie.jpg
file: adelie.jpg
size: 1.51 megabytes
key: MD5E-s1510096--96d64a3ce9491ec4a8e277bcb31bb016.jpg
present: false
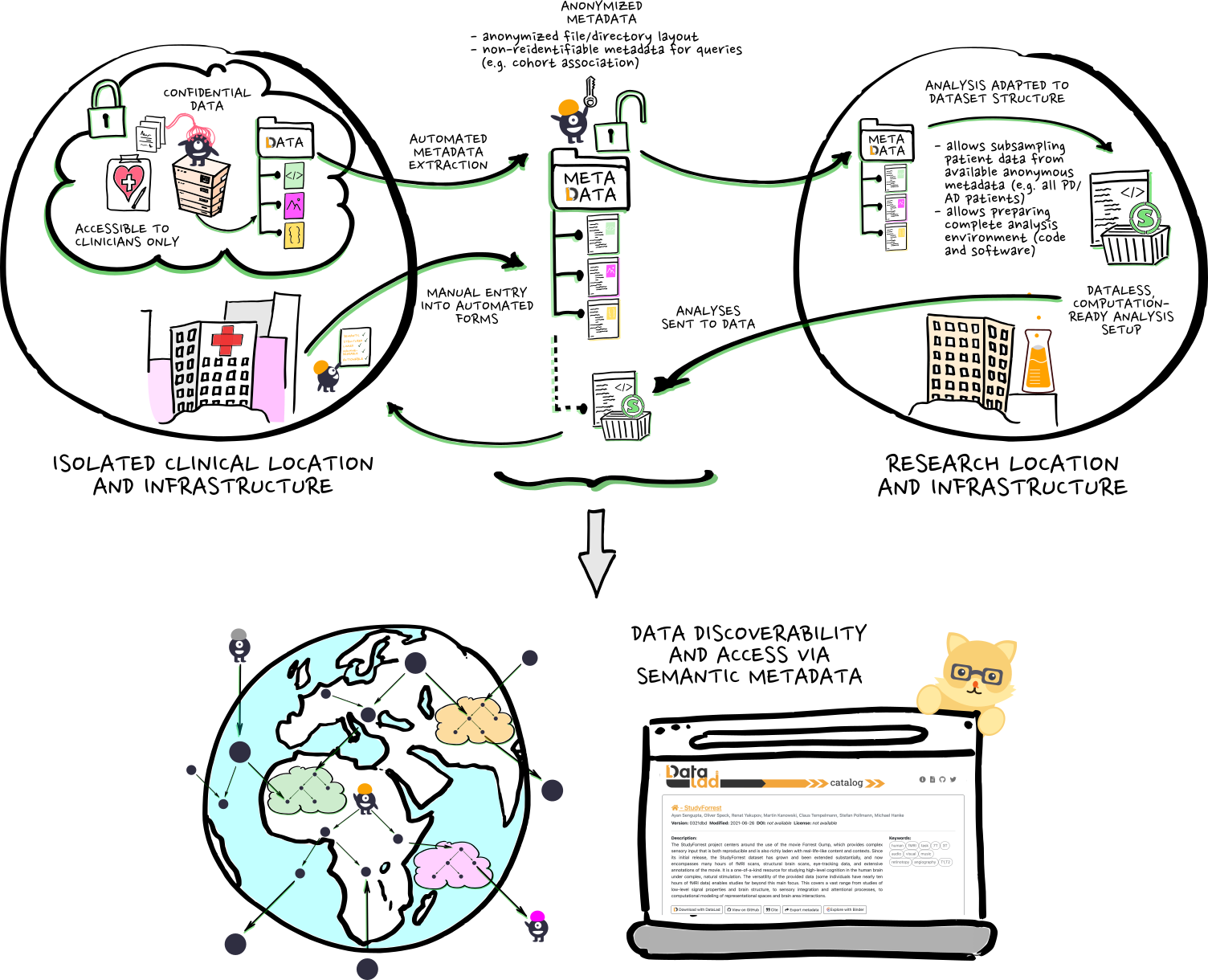
...does that make it findable?
- DataLad does not include the world
- The world should not have to care about DataLad
Human-level discoverability
A generic person will not ask:
Inside the DataLad dataset with identifier fd3b41bb-5d75-4cee-b41c-aac0e0cae7f1,
find the file with key MD5E-s18872--e4b0710c69297031d63866ce8b888f25.csv, because I walk around
with an index of git annex keys in my head.
They will ask:
In the context of the Penguin Dataset, find the data files containing the beak length of male Gentoo penguins collected on Biscoe Island during the austral summer of 2008.
=> We need to connect the inherent metadata provided by git+git-annex+DataLad (all about the files) to the broader world through contextual metadata (all about discoverability).
Connection through annotation
Let's try this out:
penguins.edu.datalad.org/ui/
For structure,
we have to model our data
we have to model our data
For structure,
we have to model our data
we have to model our data
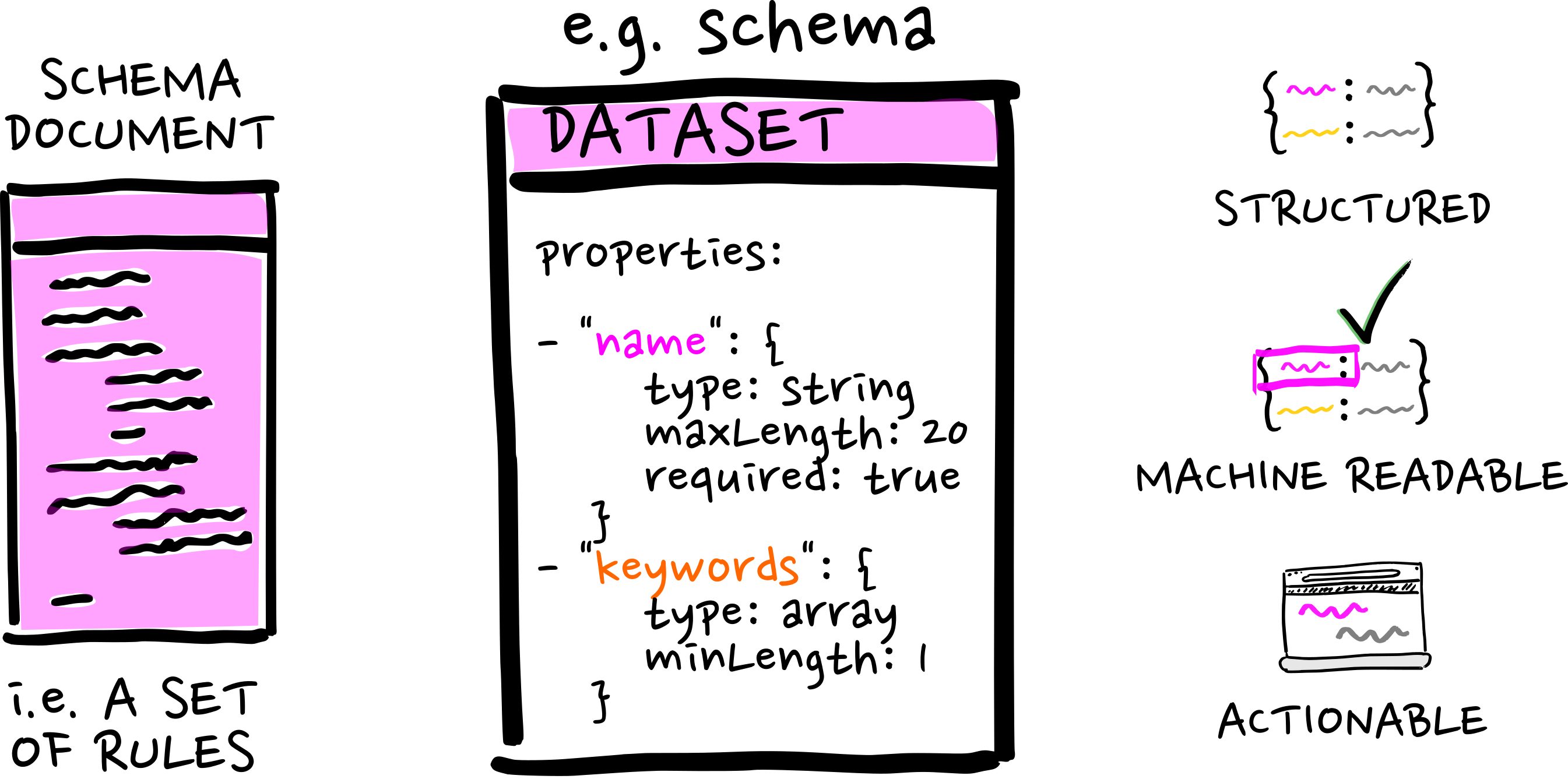
With an actionable schema...
...comes great opportunities
...comes great opportunities
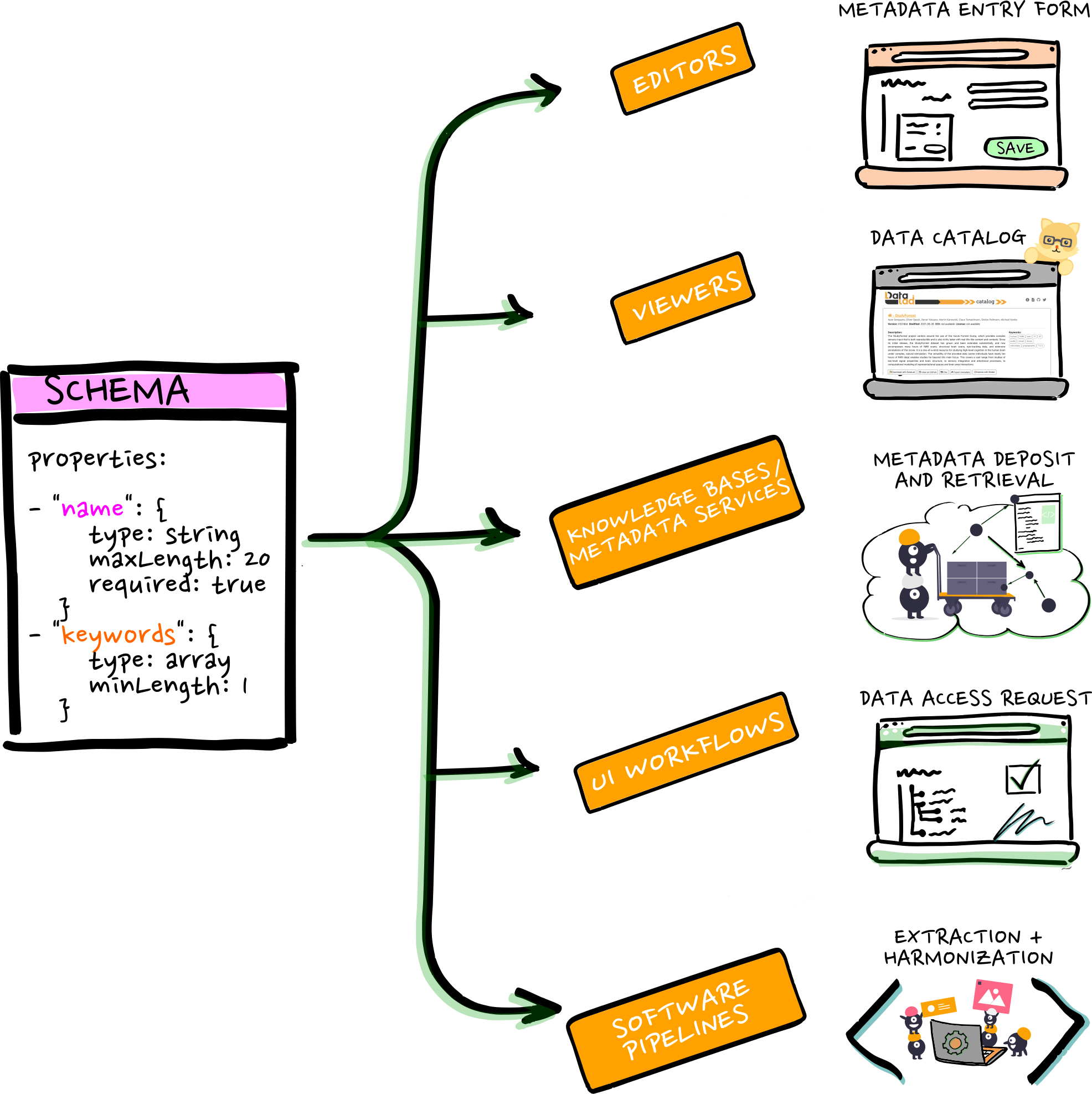
The annotation stack
Data modeling
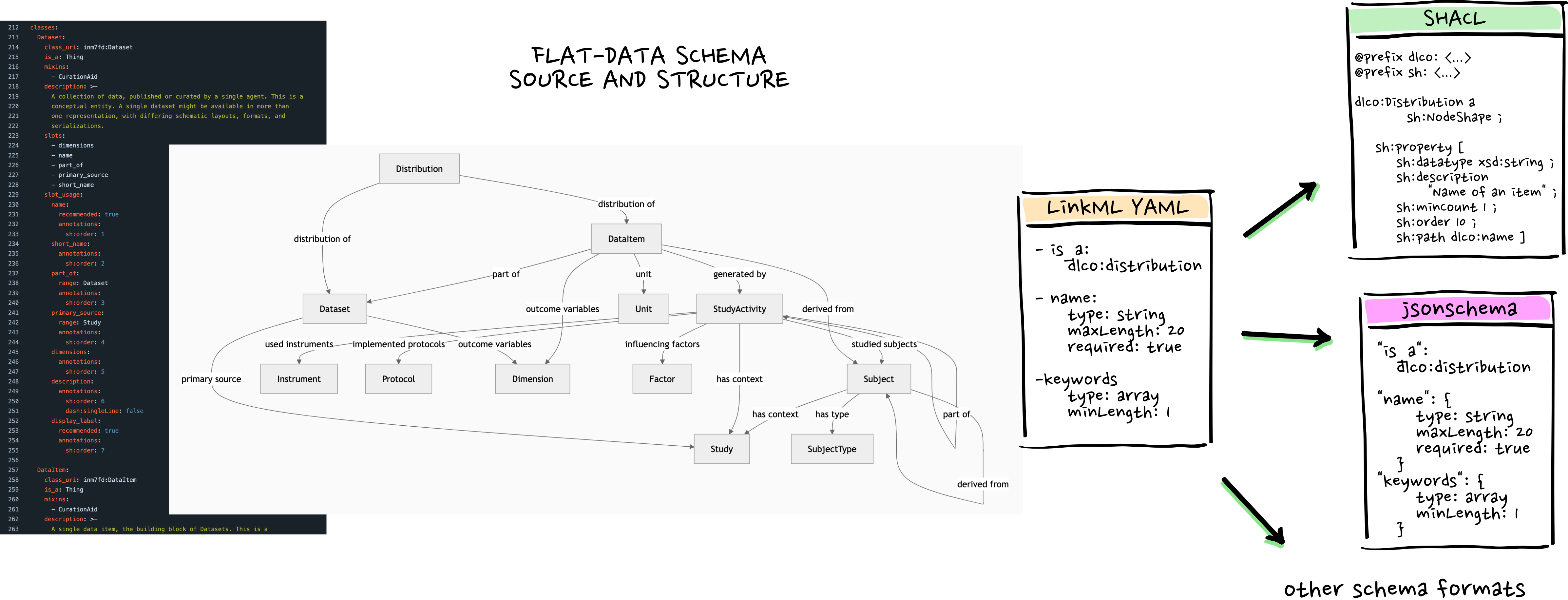
linkml.io
LinkML bridges between the worlds of structured data in plain text files, relational databases and knowledge graphs if and when needed, so metadata workflows can stay as simple as possible
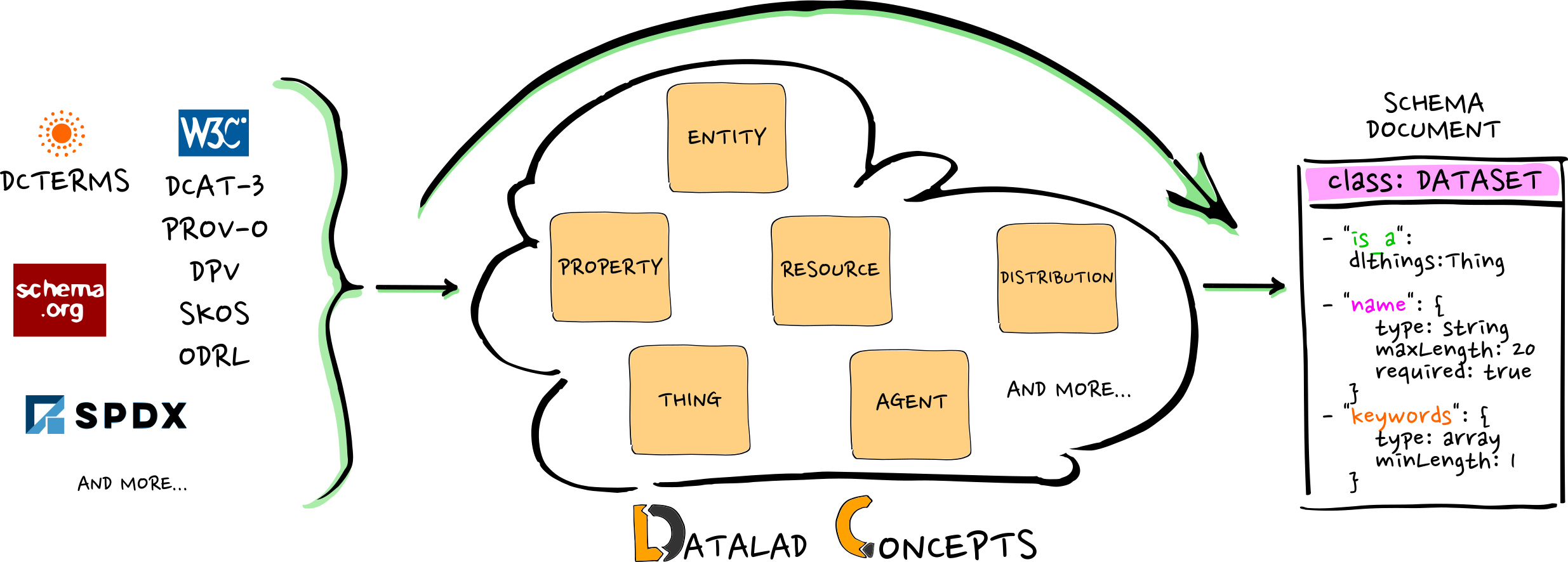
Extending semantic standards
|
- DataLad does not include the world
- The world should not have to care about DataLad
- But: DataLad is interoperable with the world!
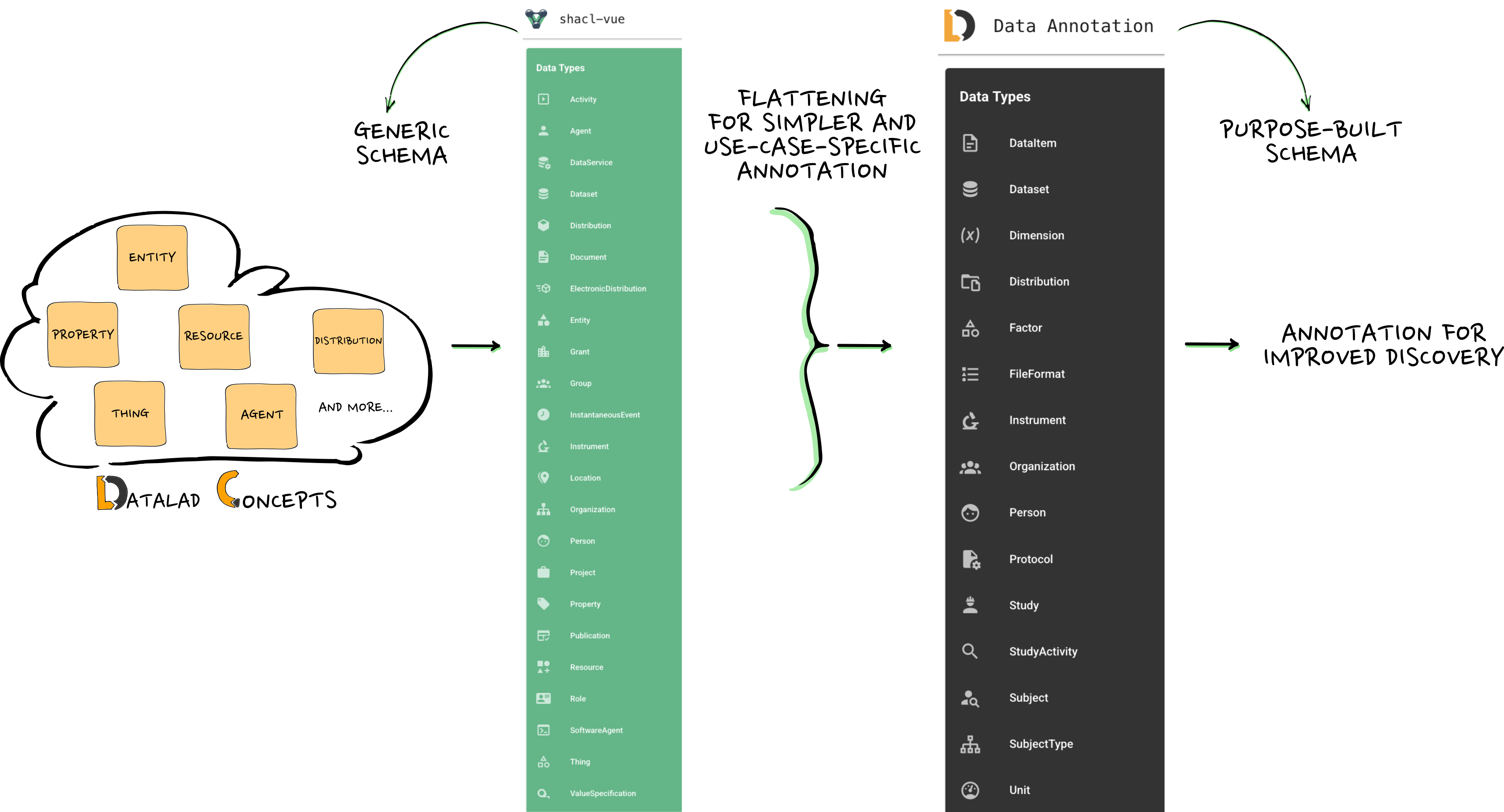
Flattening generic to purpose-built schemas
Curation and multi-schema systems
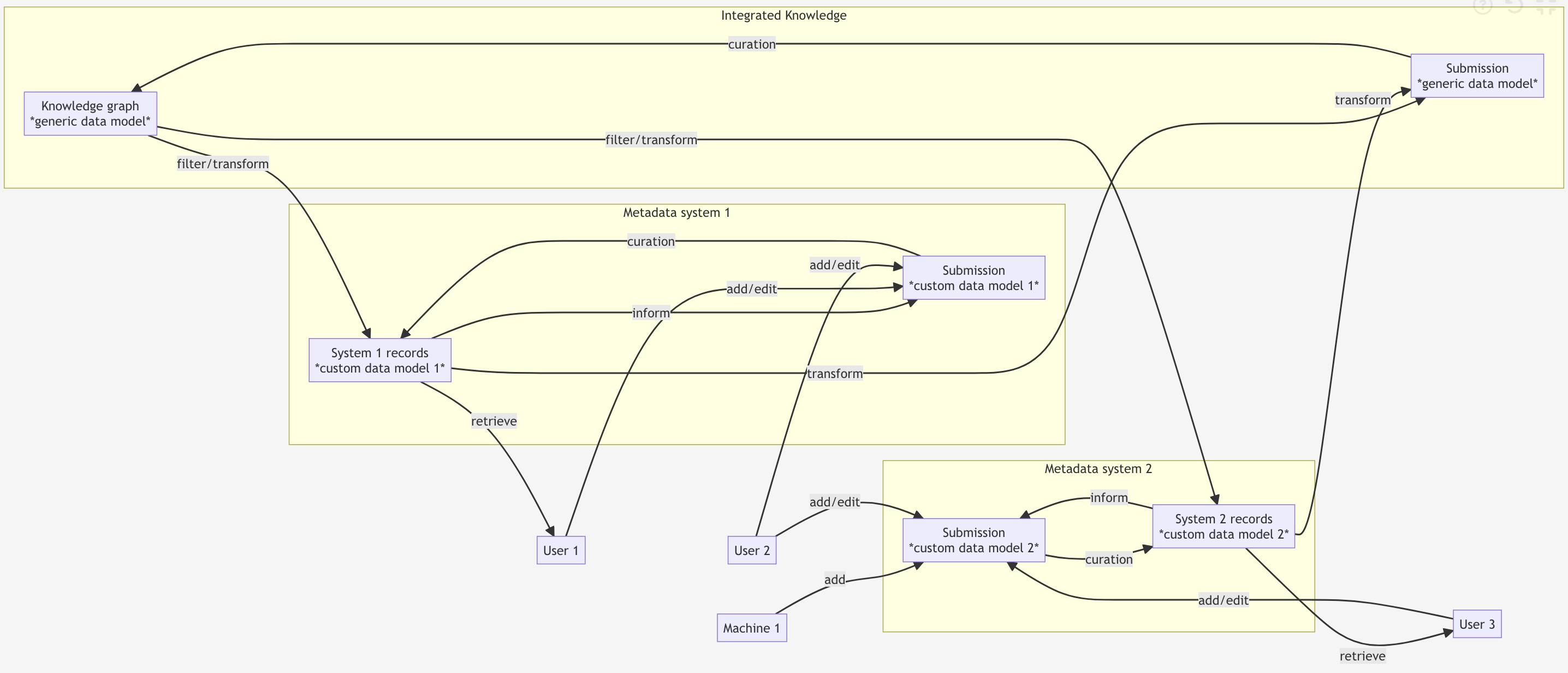
- Separate interfaces for separate purposes - all lower-level interoperable
- Staging area for curation/validation step
- Expose, share, or transform collections as needed for shared purposes: e.g.
Personfor internal personnel management AND research publications
Extra
Manual, decentralized metadata authoring
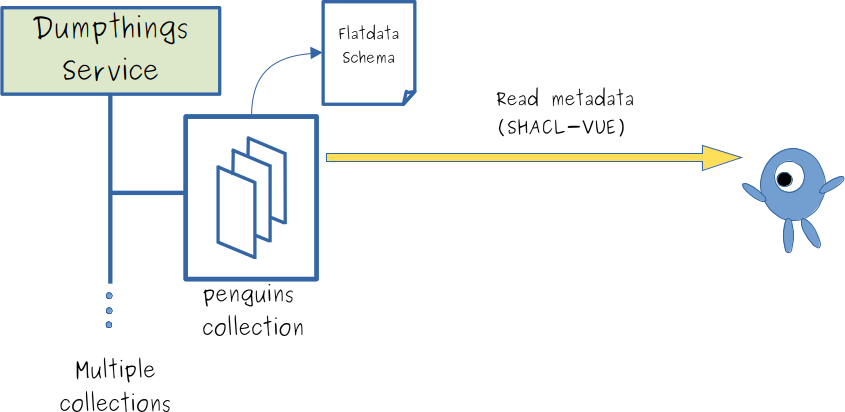
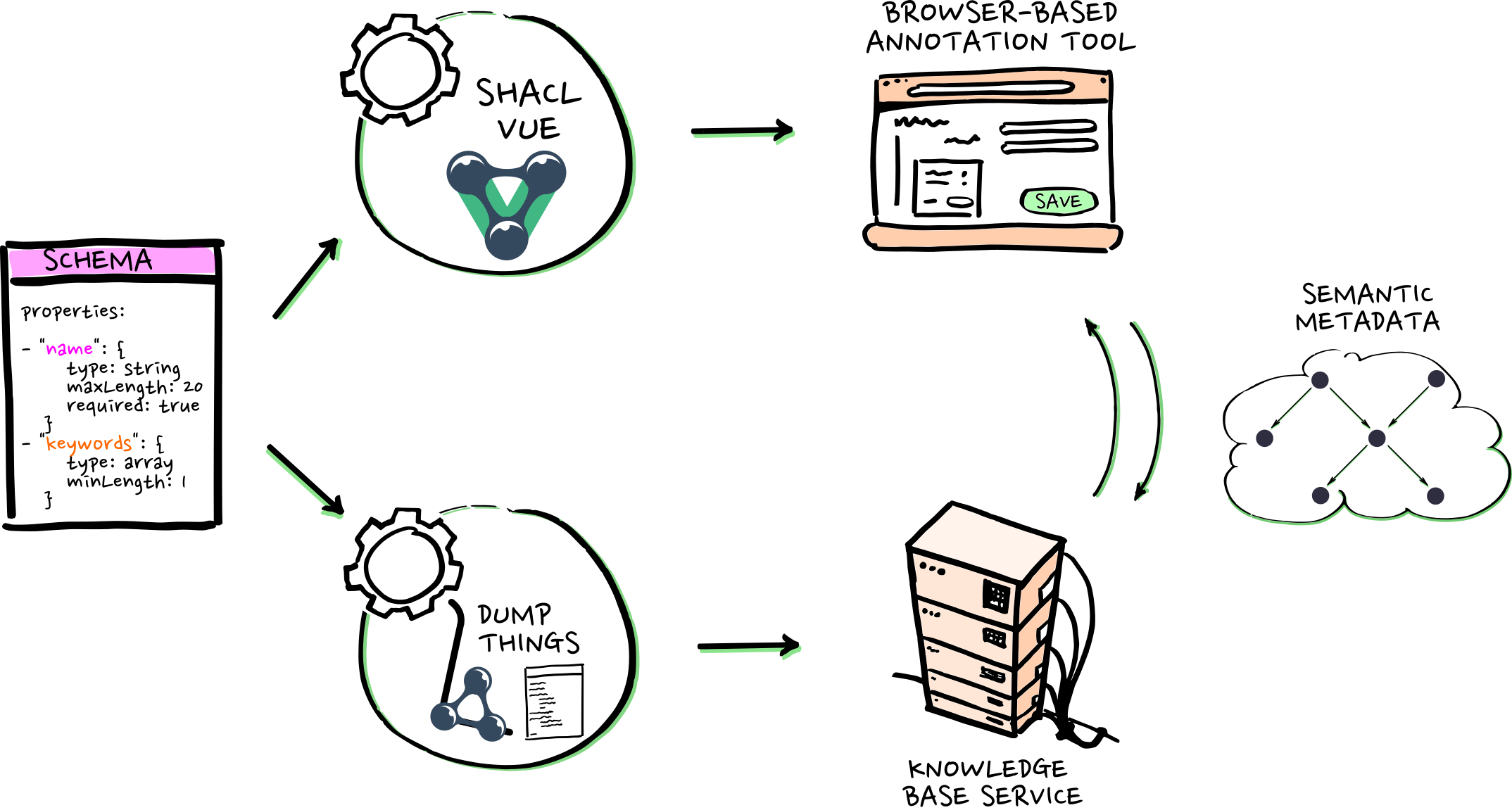
- "dumpthings"-service stores metadata in collections ...
- ... each collection has an assigned schema
- ... metadata can be read via Web-API, e.g., by SHACL-VUE
Manual, decentralized metadata authoring
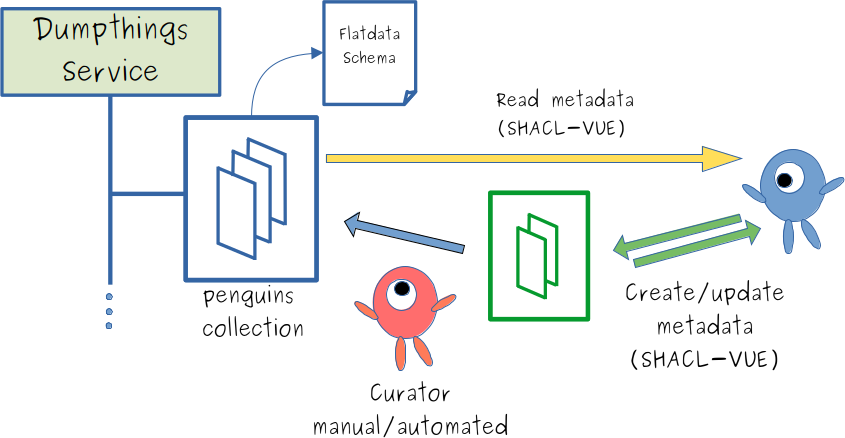
- "Users can add metadata to a collection ...
- ... to incoming space of the collection
- ... curation process moves incoming metadata to collection
Manual, decentralized metadata authoring
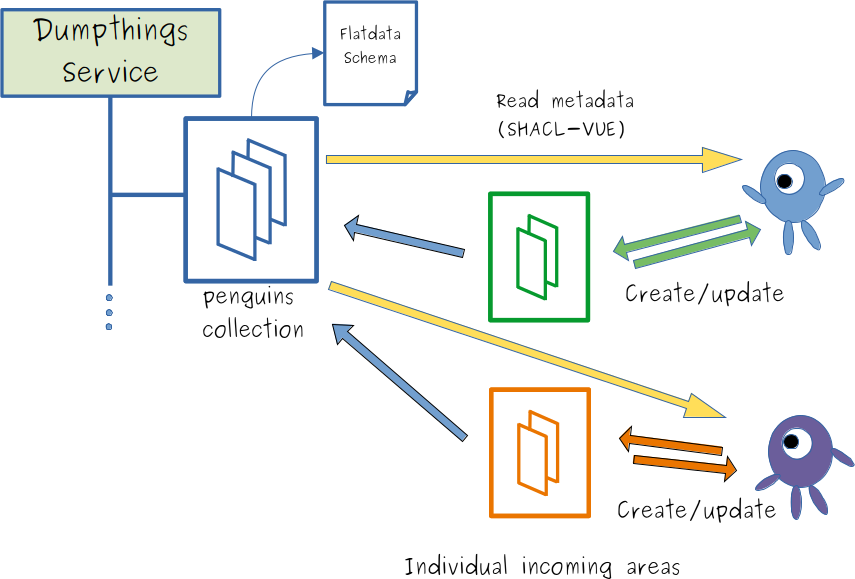
- "Multiple users can write metadata ...
- ... to individual incoming spaces
- ... shared, decentralized metadata creation
AN OPPORTUNITY:
can metadata solve this?
can metadata solve this?
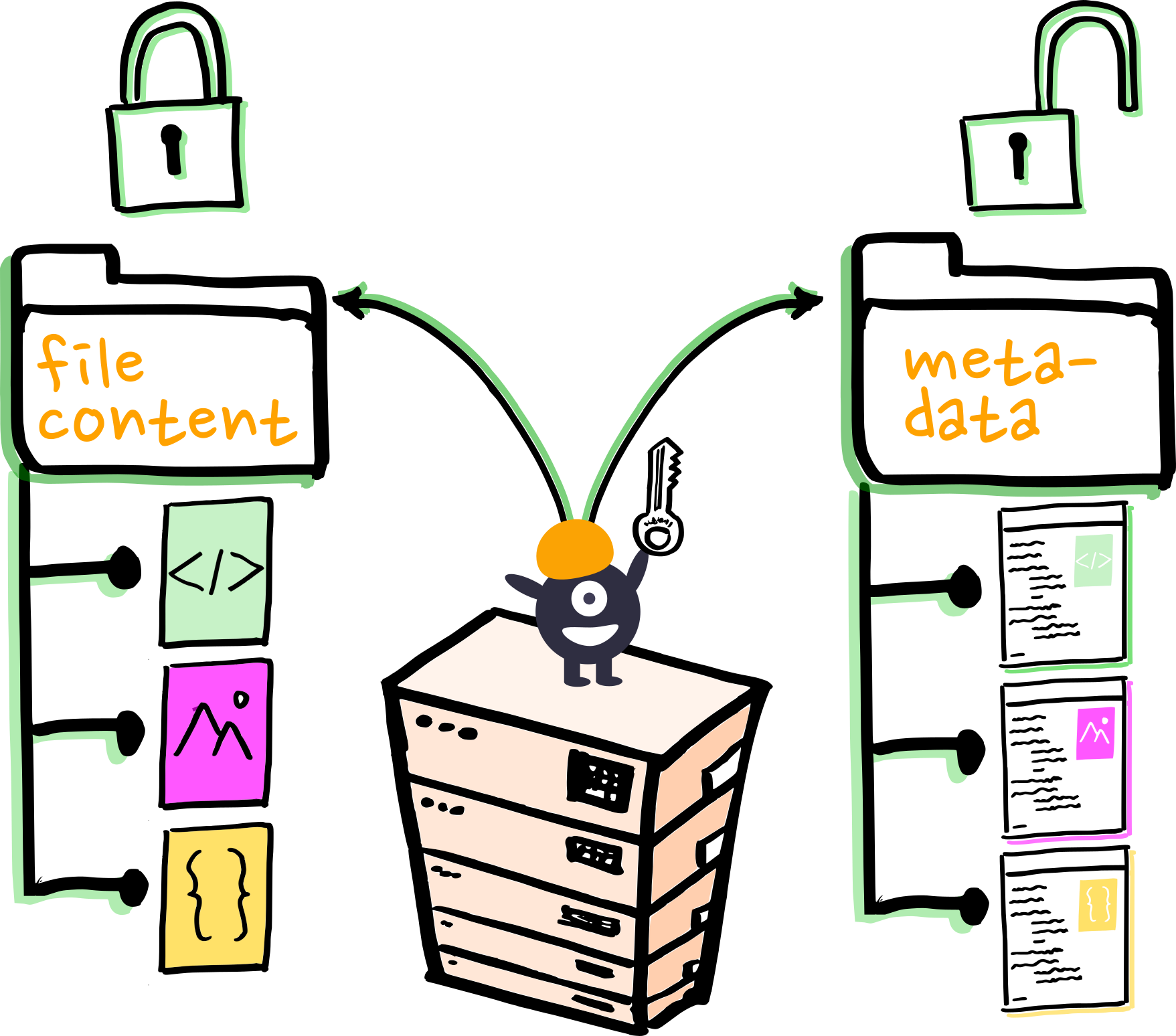
AN OPPORTUNITY:
can metadata solve this?
can metadata solve this?
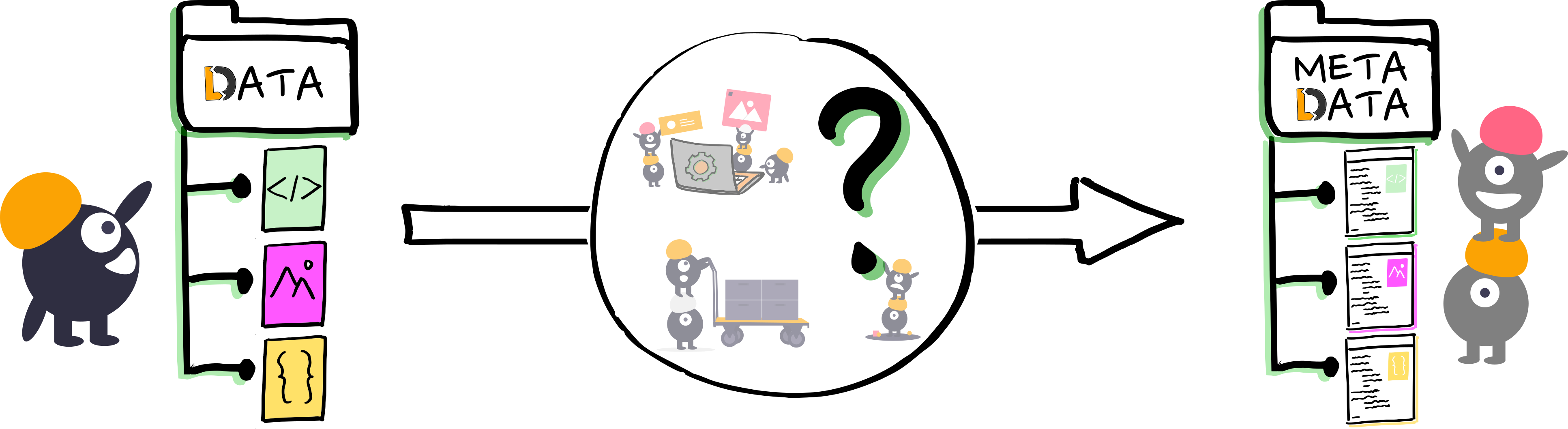
So what is metadata actually?
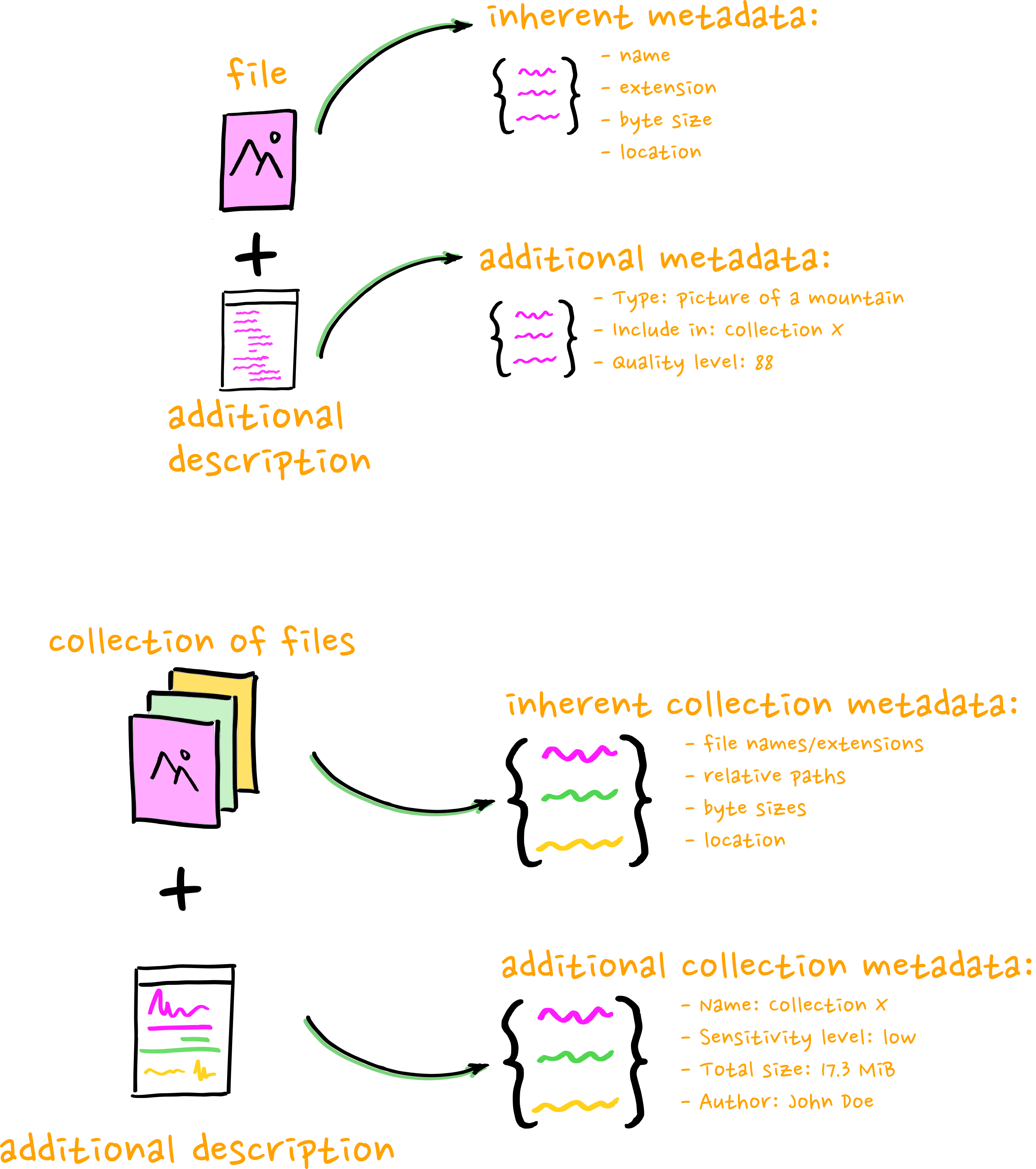
=> Inherent and contextual metadata
Where do we source inherent metadata?
Where do we source contextual metadata?
- Metadata adhering to existing standards
- Metadata entered when uploading data to a repository
- Everywhere, basically...
To be FAIR,
metadata need to be actionable
metadata need to be actionable
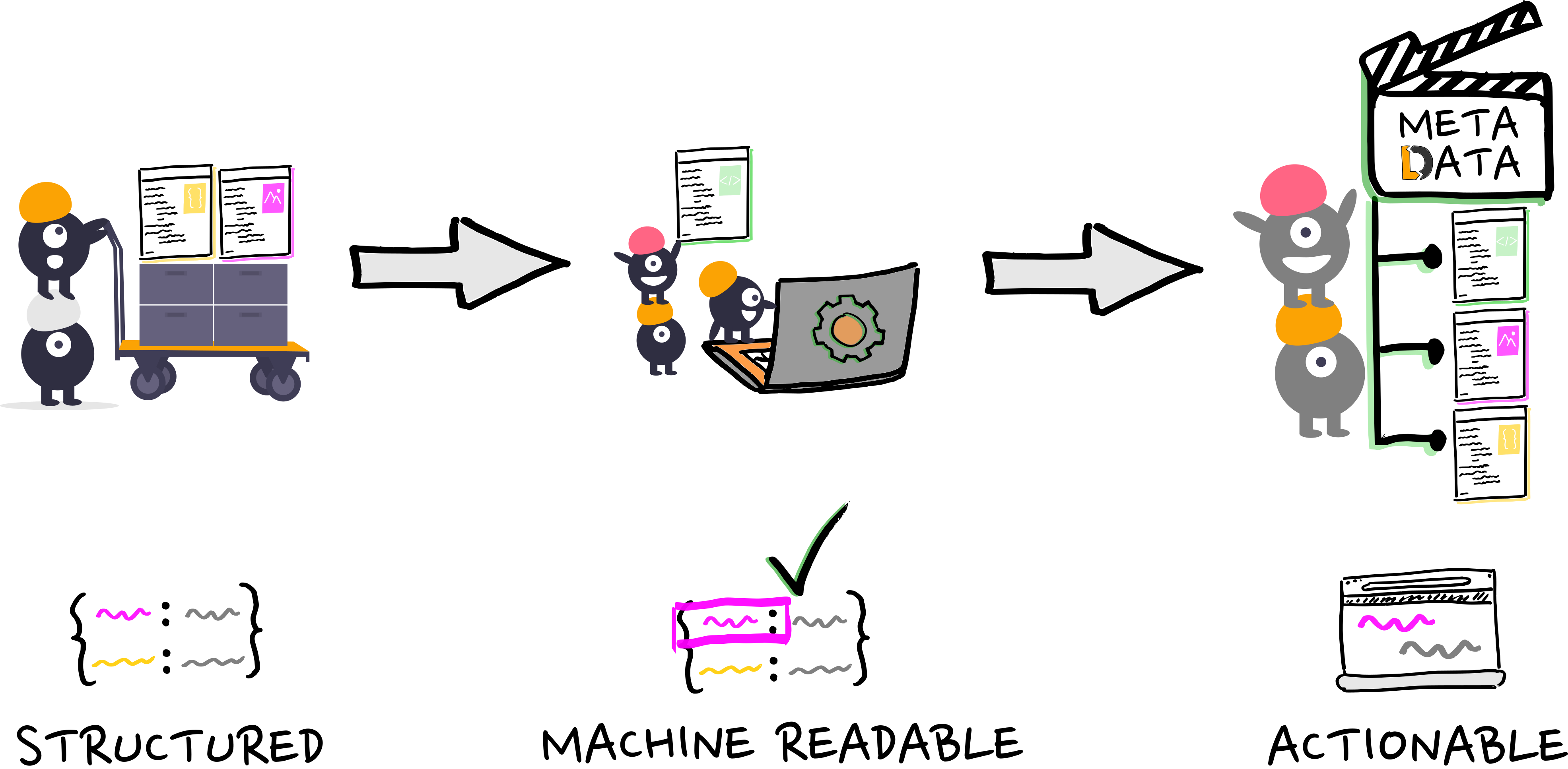
Practically, how do we get actionable metadata?
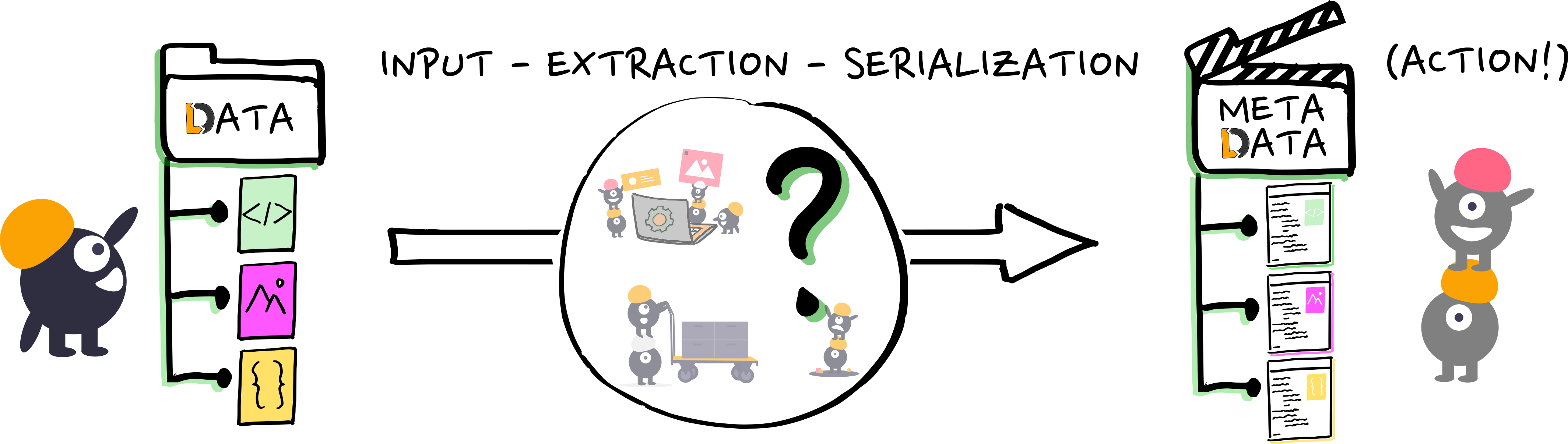
- It comes down to serialization, via...
- Researchers and software pipelines
- But first: we have to provide structure to metadata
- allows intuitive manual forms that validate on entry and that minimize effort and generality
- allows automated metadata extraction
LinkML Modeling
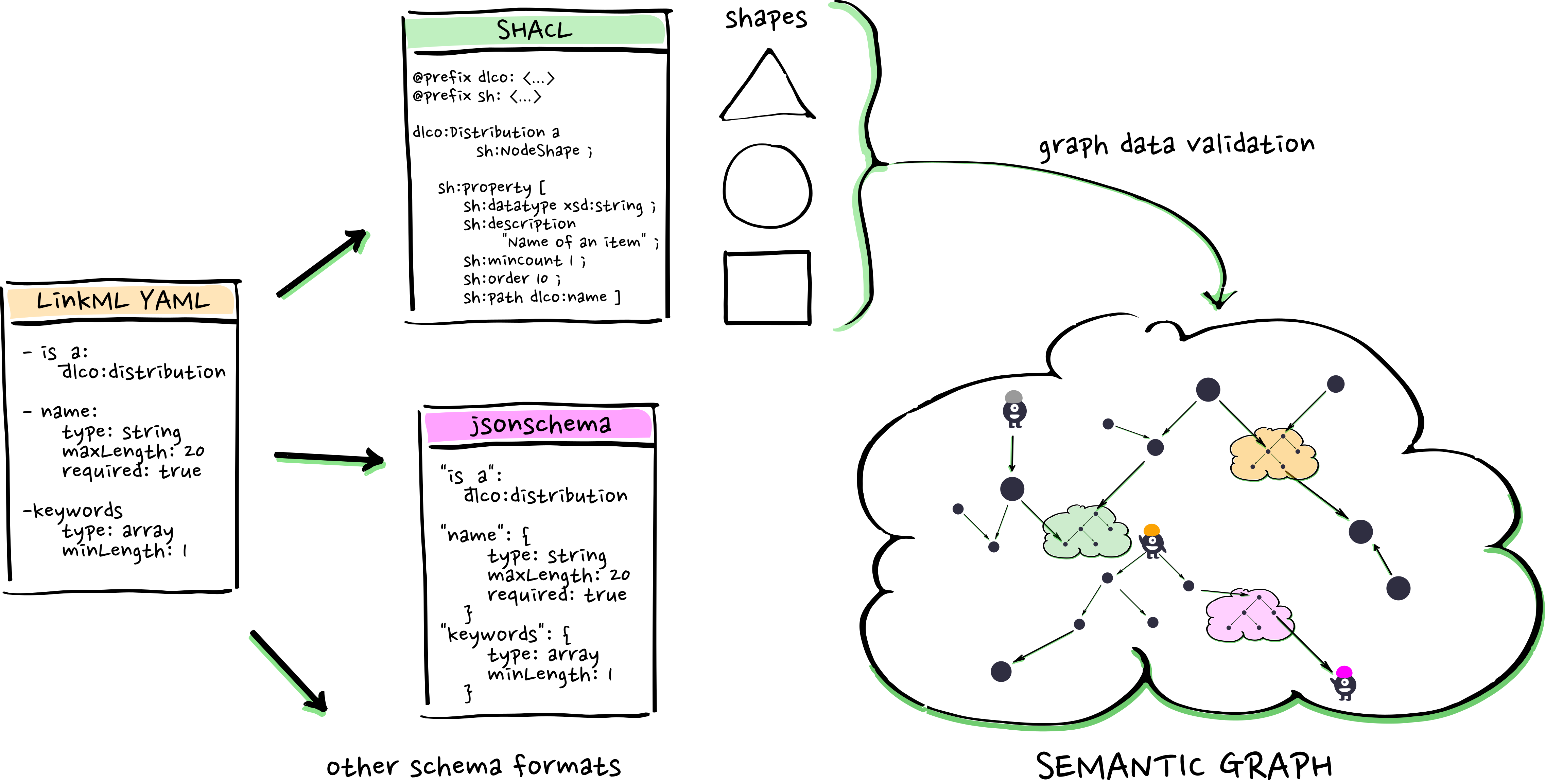
RDF, SHACL, and automation
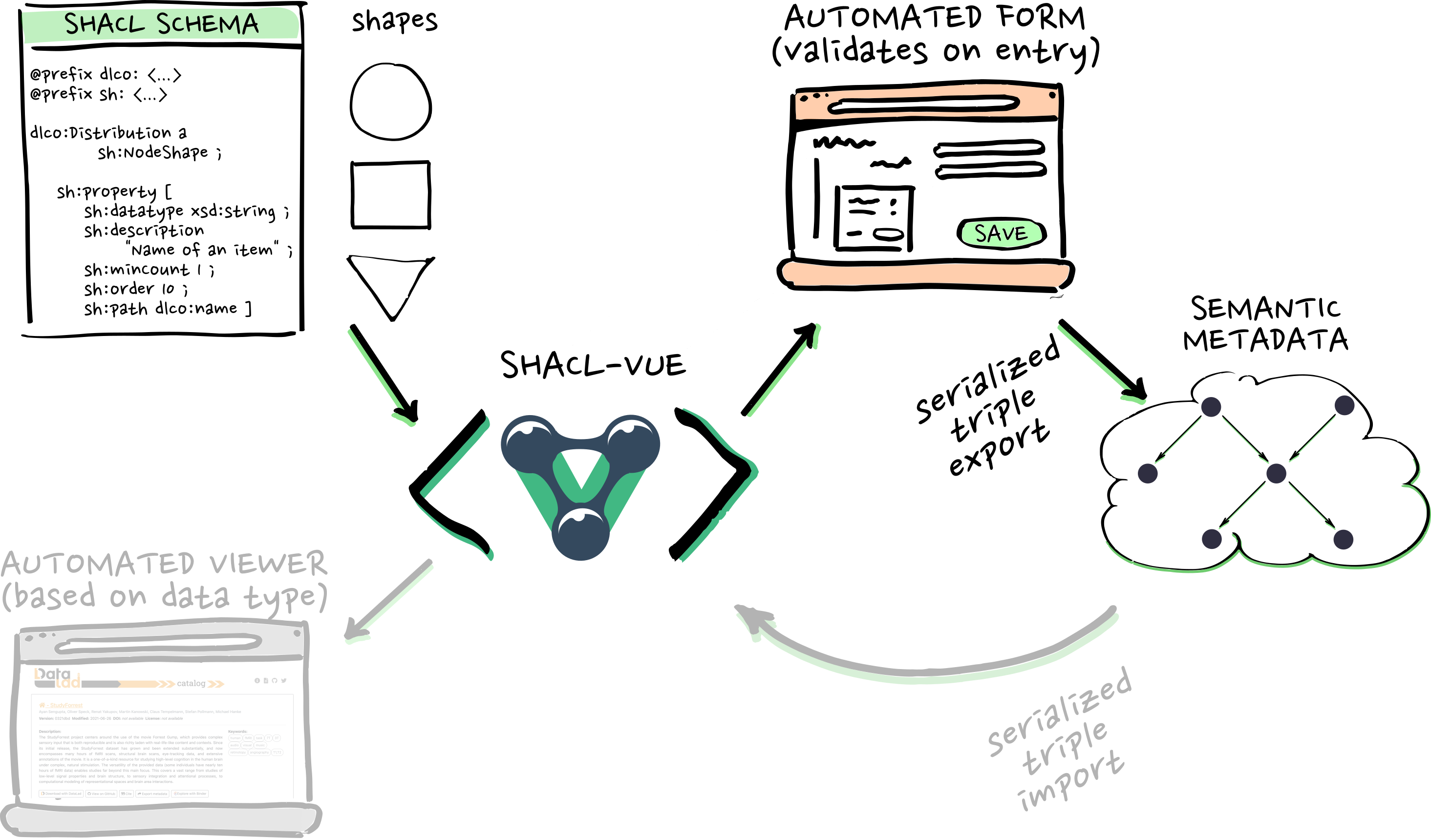
RDF, SHACL, and automation
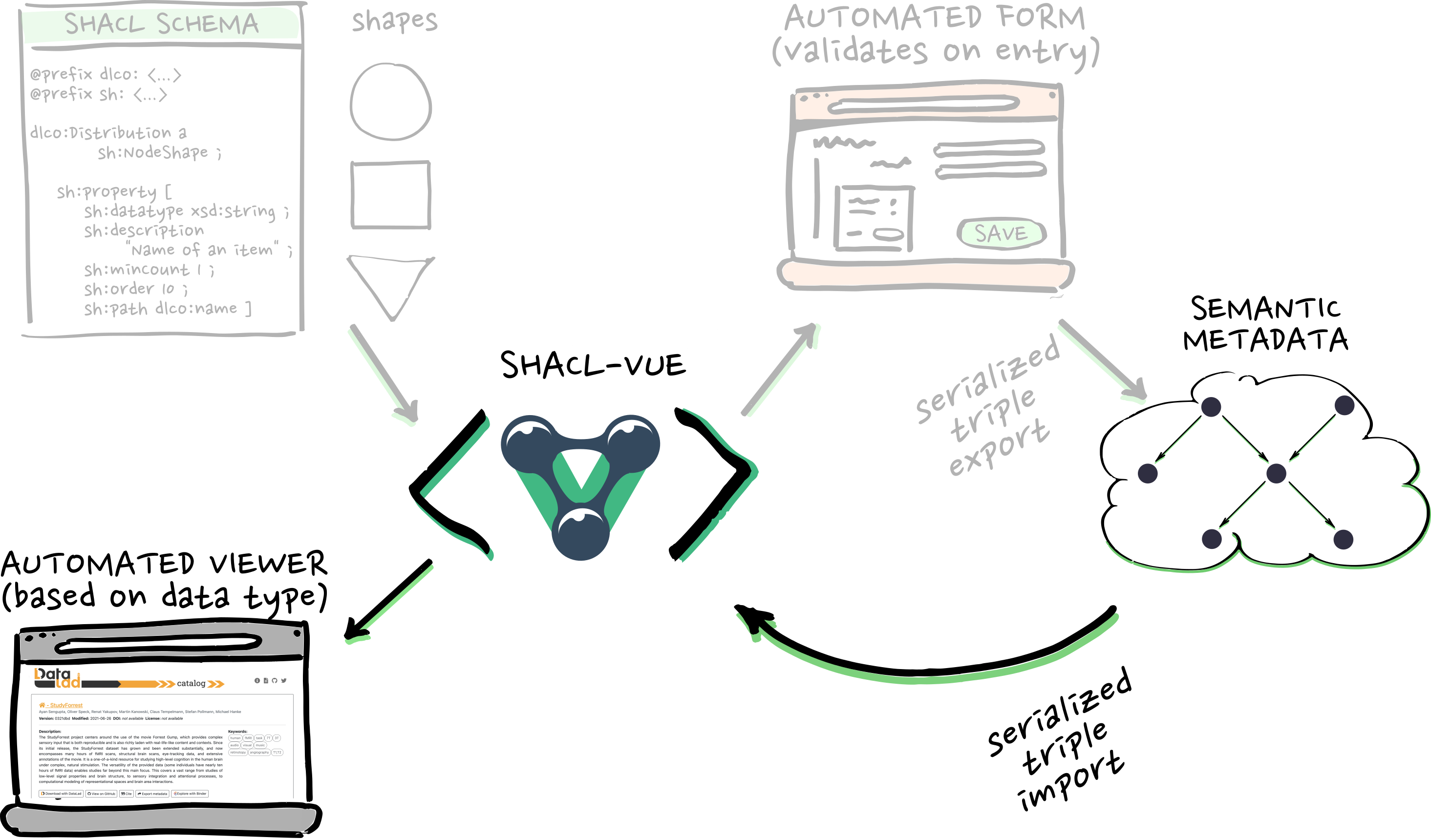
RDF, SHACL, and automation
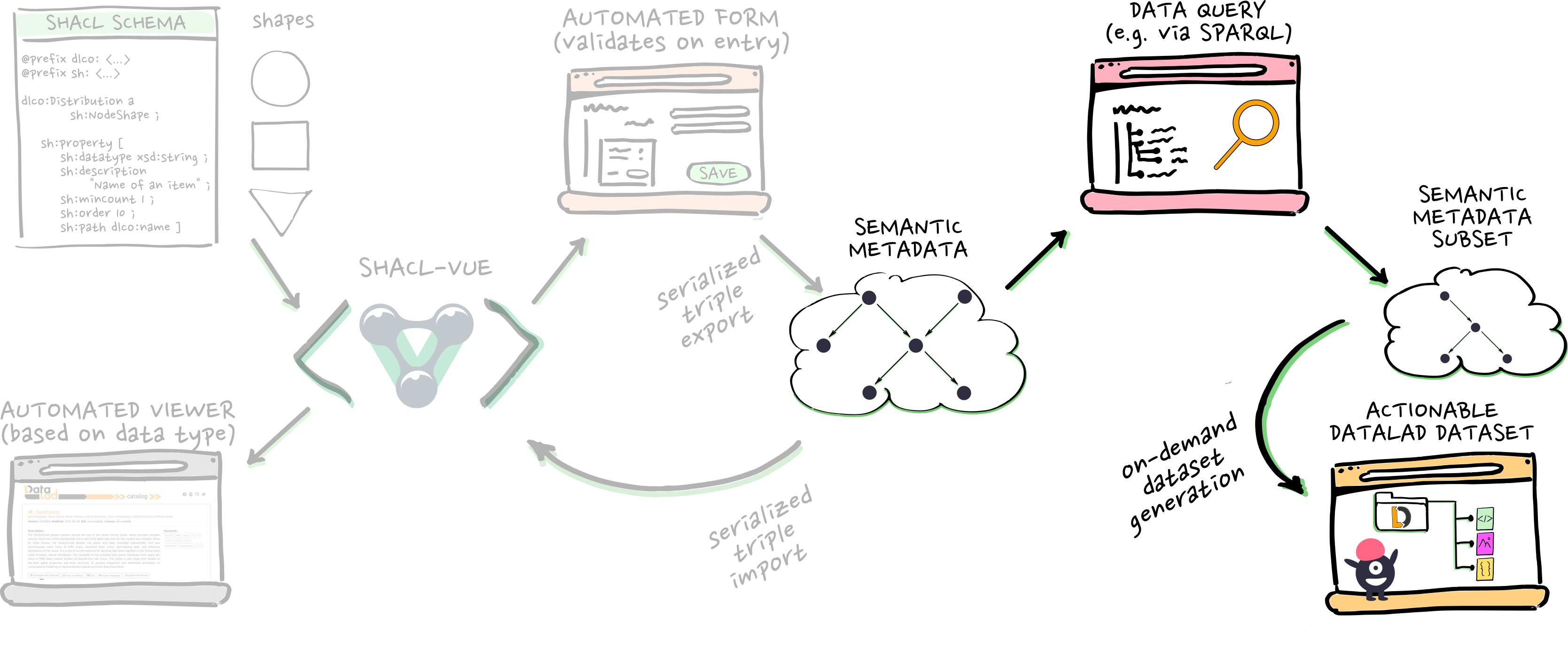
- i.e. cohort searches/queries. And then...
- ...metadata-based dataset generation, which gives us...
- ...on-demand access!
Getting back to FAIR data...